The Salmonella Foodborne Syst-OMICS Database
A Syst-OMICS approach to ensuring food safety and reducing the economic burden of Salmonellosis
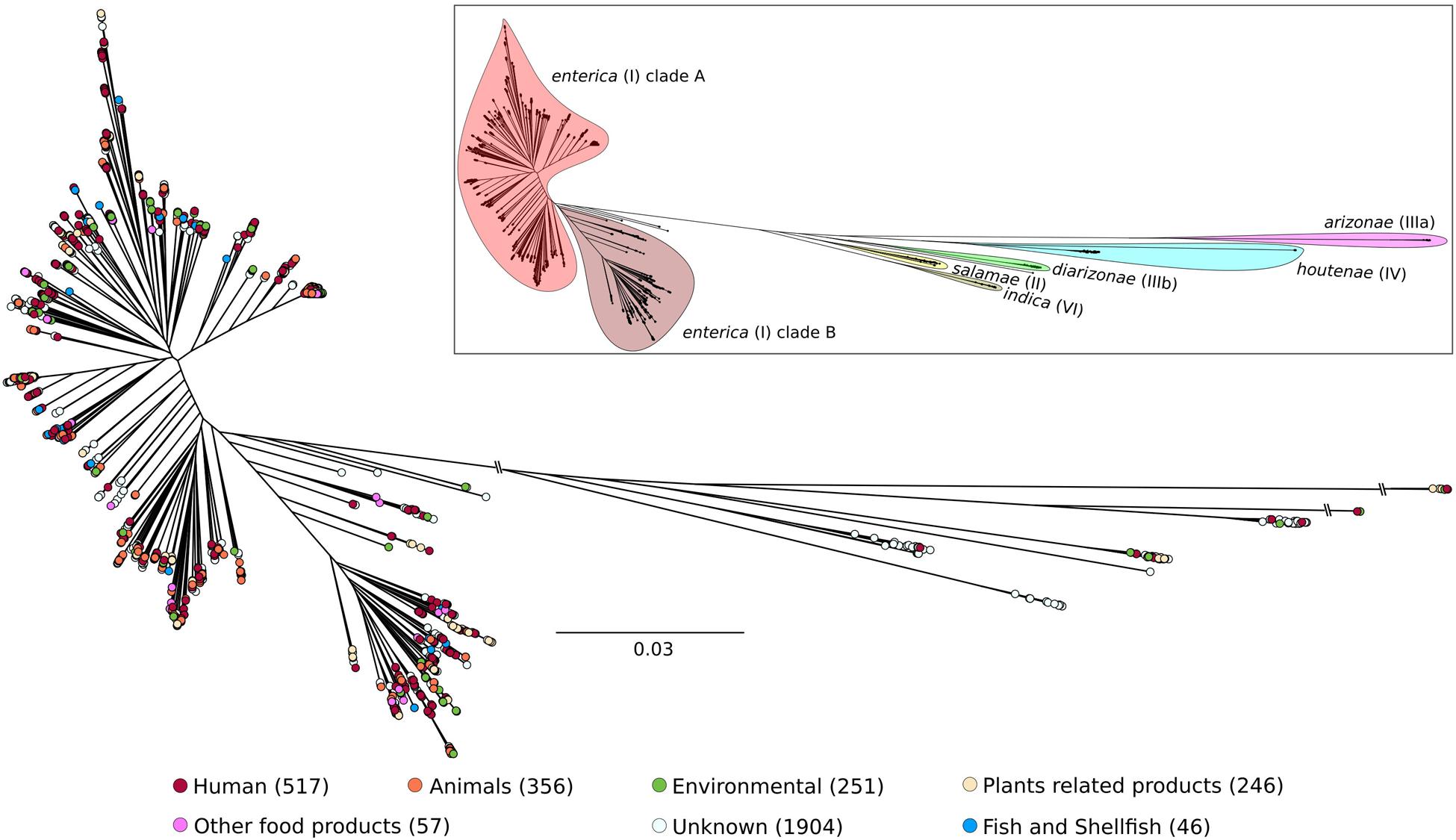
SalFoS is a repository of thousands of Salmonella isolates from the environment, plant and animal food products as well as human infections, with a strong focus on fruits and vegetables.
SalFoS currently contains 3543 isolates.
Draft genomes have been produced for 3543 of them.
The content of the SalFoS database can be accessed here
Other useful resources:
Salmonella Syst-OMICS: salmonella-systomics.ca/en/overview/
SISTR: Salmonella In Silico Typing Resource: github.com/phac-nml/sistr_cmd
Centers for Disease Control and Prevention, Salmonella: www.cdc.gov/salmonella
Salmonella enterica Pathway Genome Database in BioCyc : salmonella.biocyc.org
Enterobase: enterobase.warwick.ac.uk
PATRIC, The bacterial bioinformatics database and analysis pipeline: www.patricbrc.org
The Comprehensive Antibiotic Resistance Database (CARD): arpcard.mcmaster.ca
The Public Databases for Molecular Typing and Microbial Genome Diversity (PubMLST): pubmlst.org
Virulence Factor Databases (VFDB and Victors): www.mgc.ac.cn/VFs and phidias.us/victors/